Related article: Familial aggregation
Table of Contents
Transmission of bacteria and onset of chronic disease
Pathogens that grow slowly and accumulate over the course of decades may play a strong role in many chronic diseases. These bacteria are transmitted in a variety of ways: mother to fetus, sperm to embryo, and among families and social groups. Particular patient groups without the benefit of a fully functioning immune system, specifically newborn infants, people who already have illnesses, and the elderly, are uniquely susceptible to pathogens.
Those who use or consume any of the foods, drugs, and supplements which exert immunosuppressive effects are also uniquely predisposed to acquire new bacteria and permit them to reproduce. These substances include: immunosuppressants, beta-lactam antibiotics such as penicillin, high levels of vitamin D, and corticosteroids.
The acquisition of new bacteria is only one factor in the when and why chronic diseases strike. Bacteria are capable of rapidly changing their genetic structure – and can become more pathogenic and harder to kill with traditional therapies – through processes like horizontal gene transfer. Also, bacteria are allowed to proliferate because of a weak immune response, for which they themselves are at least partially responsible.
Vectors for transmission
It is commonly agreed upon that acute infections such as gonorrhea, influenza, and the common cold are transmitted via bodily fluids and in some cases via physical contact and breathing. Evidence is accumulating that chronic pathogens can and are transferred between people in ways previously unimagined and that these pathogens contribute to onset of chronic disease. For example, Weyermann et al. has shown that infected siblings, mothers, and fathers are all major sources for Helicobacter pylori acquisition among young children, with the infected mother likely to be the main source for childhood infection.1)
From father to child via sperm
Research indicates that pathogens are able to survive in sperm, so a father can pass these bacteria to his child at the moment of conception.2) This may explain why, according to some anecdotal reports, females who have experienced multiple miscarriages have successful pregnancies after changing partners.3) Given its key role in innate immune function, the existence of the VDRThe Vitamin D Receptor. A nuclear receptor located throughout the body that plays a key role in the innate immune response. in sperm also suggests that sperm are susceptible to infection.4) In 2012, Blomberg et al. showed that expression of the vitamin D metabolizing enzyme CYP24A1 in human sperm was an effective marker of semen quality.5)
From mother to fetus during pregnancy
Previously unrecognized, uncultivated, or difficult-to-cultivate species may play a key role in the initiation of preterm birth.
Y.W. Han 6)
A mother's womb was once considered to be sterile but studies show that chronic pathogens persist in the endometrium. Eighteen different taxa of microbes were recently identified in the amniotic fluid of women who gave birth prematurely.7) Mycobacterium tuberculosis and influenza have been shown to cross the placental barrier.8) 9) Infection with Shigella has been proposed as an explanation for the etiopathogenesis of endometriosis10) and invasion of the endometrium by bacteria has been implicated in implantation failure, spontaneous abortion, and preterm birth.11)
Evidence is also growing that certain bacteria and viruses are able to cross the placental barrier – meaning they can be passed from a pregnant woman to her fetus. Researchers now believe that the fetal gut is colonized during pregnancy. According to David Kinross: “Standard medical teaching is that the fetus is sterile. The notion that gut development is influenced by maternal bugs will come as a shock. It's a completely new way of thinking about human disease.”
The amniotic fluid of women not in labor (with intact membranes) has traditionally been considered to be free of bacteria based on studies using cultivation techniques.12) But women who deliver pre-term infants (who tend to be less healthy) has been found to contain greater numbers of bacteria than those who deliver at term. According to the authors, the data they gathered “supports a causal relationship” between the bacteria and the early pregnancies.13)
People whose parents harbor high loads of pathogens are much more likely to fall ill with a chronic disease earlier in life. Research indicates that L-form bacteriaDifficult-to-culture bacteria that lack a cell wall and are not detectable by traditional culturing processes. Sometimes referred to as cell wall deficient bacteria. are able to survive in sperm, so a father can pass these pathogens to his child at the moment of conception. Evidence is also growing that L-form bacteria and other pathogens are able to cross the placental barrier – meaning they can be passed from a pregnant woman to her fetus.
Researchers at Peking University in Beijing recently discovered that the H5N1 bird flu virus can pass through a pregnant woman’s placenta to infect her fetus.14) Other studies have revealed that other bacterial species such as Borrelia burgdorferi and Mycobacterium tuberculosis are also capable of crossing the placental barrier during pregnancy.15) If these pathogens can be passed from mother to child during gestation, then why not other forms of bacteria that are capable of transforming into the L-form?
Inflammation, the body's evolutionary conserved response to microbes, may also weaken the amniotic membrane. Kobayashi et al. applied certain cytokinesAny of various protein molecules secreted by cells of the immune system that serve to regulate the immune system. such as TNF-alphaA cytokine critical for effective immune surveillance and is required for proper proliferation and function of immune cells. to an organ-cultured amniotic membrane. This caused dysfunction of the amniotic barrier and disruption of tight junctions.16)
To newborns in their first weeks of life
A person’s age at the time of infection — from intrauterine [occurring within the uterus] or perinatal (the time period surrounding birth), through childhood and adolescence, to adulthood and the elder years — may further influence the risk for chronic outcome.
Siobhain O'Connor, et al., Centers for Disease Control and Prevention 17)
Dave Relman and team at Stanford University found that infants pick up many of the species that make up their gut flora from family members within a few weeks of birth.18) Another team found that Human papillomavirus type 16 (also called high-risk HPV-16), which has been linked to cervical cancer, can be detected in human breast milk collected during the early period after a woman delivers her baby.19)
This is evidence that pathogens are easily transmitted from family to child during the initial periods of life. Both the innate and the adaptive immune systems of infants are particularly susceptible to pathogens, which can drive chronic disease later in life.
Adaptive immune system
While the innate immune system, the body's first line of defense against pathogens, is functioning at birth, it takes several weeks for an infant to develop a working adaptive immune system. Recent research has shed light on this formative period of life.
Nobel Laureate Rolf Zinkernagel injected cytomegalovirus (CMV) into the brains of a group of newborn mice. The adaptive immune system of these mice had not yet developed and consequently they were not producing lymphocytes. The researchers found that the innate immune systems of the mice were able to eliminate CMV from most of the tissues except for those of the central nervous system. As a result, the virus persisted in the brains of the mice. Later in life, when the same mice were challenged by infection with a similar virus, they developed a condition resembling a type of autoimmune disease and died. The team referred to this concept as viral “deja vu.”20)
A second group of mice were not exposed to the CMV virus until they were fully grown and their adaptive immune systems had completely developed. When these mice were exposed to CMV later in life, they were able to successfully fight the virus and lived.
Similarly, re-exposure of aged individuals to respiratory syncytial virus (RSV) – usually the first pathogen that a human infant encounters – can cause lesions similar to those that occur in infants.21) 22)
These studies reveals what may be one of the main reasons behind why some people seem much more susceptible to the Th1 pathogensThe community of bacterial pathogens which cause chronic inflammatory disease - one which almost certainly includes multiple species and bacterial forms. (L-form, intracellular and biofilm A structured community of microorganisms encapsulated within a self-developed protective matrix and living together. bacteria) that cause chronic disease and acquire these chronic bacterial forms at higher rates than much of the rest of the population.
In genetically susceptible individuals, early childhood infections seem to predispose them to [such disease as] multiple sclerosis or type 1 diabetes years even decades before clinical onset.
Rolf Zinkernagel, MD, PhD
When assessing Zinkernagel’s study, it must be noted the human adaptive immune system takes even longer to develop than the rat adaptive immune system. While it takes rats only days to fully acquire innate immunityThe body's first line of defense against intracellular and other pathogens. According to the Marshall Pathogenesis the innate immune system becomes disabled as patients develop chronic disease., it takes humans several weeks. Certainly this model hints at why Th1 diseasesThe chronic inflammatory diseases caused by bacterial pathogens. often run in families. If parents, grandparents and relatives with high loads of the Th1 pathogens hold and care for babies during the first weeks of life, it appears that their bacteria can easily be transmitted to the child and persist in the child’s brain and other tissues.
Unfortunately, the idea of pathogen déjà vu and the presence of the Th1 pathogens are not being considered by the vast majority of other researchers and doctors besides Trevor Marshall.
The really important thing to be drawn from [Rolf Zinkernagel's] work is the reminder that the infant is unprotected in the days and weeks following birth, until it starts producing antibodies. The innate immune system is all it has got although many think that antibody transfer from the mother may be present in the breast milk. I am not sure that would be much help, but I just don’t know. Nobody does.
Trevor Marshall, PhD
Innate immune system
The infant's innate immune defense has developed various mechanisms for protecting the body against infection. For example, the vernix caseosa, also referred to as vernix, is a fatty, milky looking substance that covers the fetus beginning around the end of the second trimester and through birth. A number of antimicrobial peptidesBody’s naturally produced broad-spectrum antibacterials which target pathogens. and proteins has been identified in vernix such as the antimicrobial peptide, LL-37 (cathelicidin Family of antimicrobial peptides found primarily in immune cells and transcribed by the Vitamin D Receptor.).23) Vernix has been shown to have a wider range of activities that protect the fetus and newborn against infection including antifungal activity, opsonizing capacity, protease inhibition and parasite inactivation.24)
However, the defense is imperfect. As discussed above, some neonates' innate immune system may not be able to clear bacteria from other tissues besides the brain during the first weeks of life.
One five-year study found that newborns who harbor certain types of bacteria in their throats, including Streptococcus pneumoniae, a common cause of pneumonia, and Haemophilus influenzae, which causes upper respiratory infections, are at significantly increased risk for developing recurrent wheeze and asthma early in life.25)
According to Dr. Erika von Mutius, from University Children’s Hospital in Munich, Germany, these findings may be interpreted to suggest that the presence and growth of bacteria in the throat in the first four weeks of life “indicates a defective innate immune responseThe body's first line of defense against intracellular and other pathogens. According to the Marshall Pathogenesis the innate immune system becomes disabled as patients develop chronic disease. very early in life, which promotes the development of asthma.”26)
This finding “opens new perspectives for the understanding and prediction of recurrent wheeze and asthma in young children”, says lead researcher Hans Bisgard.
Familial contact
Spouses have a significantly greater chance of developing the same disease as their partners - a phenomenon that can best be explained if familial aggregation has an infectious cause.
- sarcoidosis among spouses – One study of sarcoidosis found that among the 215 study participants who had been diagnosed with sarcoidosis, there were five husband-and-wife couples that both had the disease, an incidence 1,000 times greater than could be expected by chance.27)
- hypertension among spouses – British researchers found that men whose spouses had hypertension had a two-fold increased risk of hypertension. Similarly, women whose spouses had hypertension also doubled their risk of developing the disease. The risk for both male and female subjects persisted after adjustment for other variables such as diet.28)
- dementia among caregivers – A subject whose spouse experienced incident dementia onset had a six times greater risk for incident dementia as subjects whose spouses were dementia free.29)
Social contact
A number of studies of unrelated people shows that mere proximity seems to be enough to transmit chronic disease. A case-controlled study of residents of the Isle of Man found that 40% of people with sarcoidosis had been in social contact with a person known to have the disease, compared with 1-2% of the control subjects.30) Another study reported three cases of sarcoidosis among ten firefighters who apprenticed together.31)
Research also suggests that the inflammatory disease obesity is caused by certain species of the Th1 pathogens. A study recently published in the New England Journal of Medicine found that a person’s risk of becoming obese increases by 57% if they have a friend who becomes obese, and by 37% if their spouse becomes obese.32) According to the researchers: “These clusters did not appear to be solely attributable to the selective formation of social ties among obese persons.”
Food
Pasteurization and refrigeration
According to Cornell researchers, milk undergoes heat treatment known as pasteurization to kill off microbes that can cause food spoilage and disease, but the bacterial strain Paenibacillus can survive this heat shock as spores and cause milk to curdle in storage.33) As spores, the bacteria can survive in dormant form for years despite the best practices in cleaning, processing and packaging. In fact, the bacteria may be uniquely adapted to overcome the twin tactics of dairy protection: pasteurization followed by refrigeration.
The spores are not only resistant to heat, the small jolt of heat during pasteurization may actually stimulate them to germinate. Some can reproduce in refrigerated dairy products at temperatures that would stymy other types of bacteria. Paenibacillus is ubiquitous in nature and cause off-flavors in a variety of foods and curdling in dairy products.
Globalization of food
Vitamin D, and possibly other medically well-intentioned interventions (like steroid asthma inhalers for kids) are certainly a factor. But the globalization of food is the biggest issue, IMO. For example, meat is exported all over the world, a major carrier of microbes. Similarly for grains.
Birds have been around for a very long time, as have penguins, and Borrelia were recently isolated from Egyptian mummies. So in evolutionary terms, in my opinion, none of the things cited (ticks, birds, penguins) are likely to have been responsible for the surge in chronic disease these last few decades. In my opinion, the answer is lying right under our noses. Perhaps literally, since our mouths could be said to be “under our noses.”
Trevor Marshall, PhD
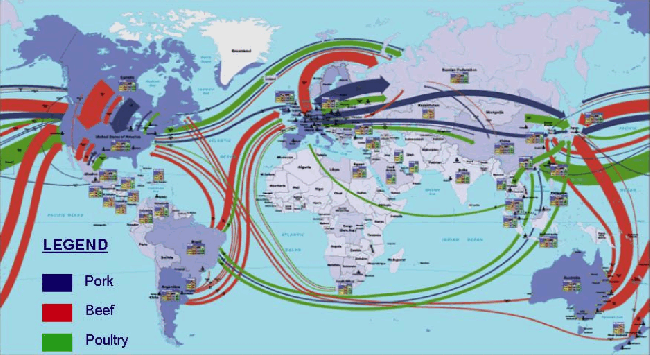
The following chart visually represents how chicken, beef, and poultry are traded on a global scale.
Microbes in meat
A study published in BMC Microbiology has looked at the prevalence of Campylobacter in skinless, boneless chicken breasts, tenderloins, and thighs. The meat was purchased in food stores in Alabama from 2005 to 2011. Campylobacter bacteria was found in 41% of the meat samples. This study reinforces the fact that consumers should avoid cross-contamination with raw poultry and should cook chicken to well-done.34) C. coli and C. jejuni had an average prevalence of 28% and 66%, respectively. The prevalence of Campylobacter did not change during the seven years of the study.
According to a 2011 research study, meat commonly found on grocery store shelves contain high levels of the microbes Staphylococcus aureus.35) The study found that in 96 percent of the meats with staph bacteria the bacteria were resistant to at least one type of antibiotic, and 52 percent were resistant to three or more types.
“Staph causes hundreds of thousands of infections in the United States every year,” the lead researcher, Dr. Lance Price, said in an interview about the study. “It causes a whole slew of infections ranging from skin infections to really bad respiratory infections like pneumonia.” He went on to say that staph infections kill more people in the United States each year than HIV.
Drinking water
Pinto et al. performed 16s rRNA gene based pyrsosequencing on water at a variety of stages in a drinking water distribution system. The team found a consistent diversity of bacteria at every stage of the multi-stage process including 16 phyla in the disinfection tank.36) Confined and unconfined aquifer ecosystems also provide conditions for growth of unique microbial communities.37)
Travel
According to a 2012 BMJ paper, elite athletes travelling to international destinations >5 time zone differences from their home country have a 2–3-fold increased risk of illness.38) Air travel does not seem to play a part as on returning home the competitors' health does not differ from normal. The researchers argue the different germs and allergens of a new environment were the key factor affecting the athletes.
In its 2017 Global Market Forecast, Airbus estimated that world air travel will grow at 4.4 per cent annually, with some 35,000 new passengers.
Injectable medicine, vaccines, and procedures
Related article: Vaccines and TB tests
Various vaccines have been shown to be contaminated and contribute to the acute onset of chronic disease.
- Guillain-Barre outbreak in 1976 – It is widely accepted that the influenza vaccine was responsible for the outbreak of Guillain-Barre syndrome in the United States in the seventies.39) In fact, in 1976, the national swine influenza vaccination program in the United States was temporarily suspended.40)
An unexplained increase in the risk of Guillain-Barre syndrome (GBS) occurred among recipients of the swine influenza vaccine in 1976-1977. Guillain-Barre syndrome remains the most frequent neurological condition reported after influenza vaccination to the Vaccine Adverse Events Reporting System (VAERS) since its inception in 1990.
Penina Haber, et al. 41)
- Clinical trial for AIDS vaccine – A 2007 international trial for an AIDS vaccine had to be abruptly discontinued when scientists realized that the vaccine somehow raised the risk of infection. The researchers administering the trial stressed that the vaccine could not itself cause the infection. Although it was not mentioned in the news story about the incident, one plausible hypothesis is that the vaccines were infected with bacteria.
The literature contains multiple reports of sarcoidosis patients developing skin lesions within tattoos. According to one researcher, this is “a well-recognized occurrence in patients with sarcoidosis.”47) One case report describes how a patient developed sarcoidal granulomas in only one pigment of a tattoo,48) suggesting, of course, that the needle corresponding to that pigment was infected.
That a tattoo procedure could routinely induce this kind of reaction strongly suggests that some needles are infected with bacteria.
Donated blood and body tissue
Donation of blood, bone marrow, organs or other tissues transmits pathogens between donor and recipient - even, in the case of blood donation, when there is an attempt to filter blood. At an average of around 0.01 microns in diameter, L-form bacteria are small enough to pass through even the finest of filters.49)
Organs and tissue from sarcoidosis patients have been known to cause sarcoidosis in the transplant recipients.50) According to one study, patients who receive a donor organ from a sarcoidosis patient develop the disease, and clean organs transplanted into sarcoidosis patients become infected.51) This is proof that the bacterial pathogens can be transferred and trigger the same abnormal immune system response in susceptible people.
Insect bites and other infectious triggers
As with mosquitoes, which transfer malaria in tropical areas, insect bites including ticks can transfer pathogenic bacteria. Chung et al found that a cancer drug, Erbitux, was more likely to cause an allergic-type reaction in regions of the country with higher insect populations.52)
It is widely assumed that chronic Lyme disease is the result of a tick bite and occurs when bacteria such as Borellia burgdorferi is transferred to a human host.53) 54) However, this explanation is incomplete. Only about 5% of people who are bitten by a tick and fall ill with Lyme disease go on to develop chronic Lyme disease, a condition where the body is simply unable to kill the bacteria responsible for causing the illness. It is theorized that the pathogens which cause Lyme disease are able to persist by transforming into the L-form and living inside the tissues and the cells of the immune system.
As discussed above, Zinkernagel and Bisgard have shown that exposure to a pathogen at a formative time in development can later predispose an individual to chronic disease, a phenomenon Zinkernagel calls “bacterial déjà vu.” In the context of chronic Lyme disease and any of the other diseases thought to be caused by insect bites alone, bacterial déjà vu makes sense. An infant is exposed to a certain pathogen early in life when the immune system is not fully functional, and becomes uniquely predisposed him or her to triggers for chronic illness later in life.
A case report
Matt Russell used the MP to treat his chronic Lyme disease. Matt was born five weeks premature. He was breast-fed, but also given formula supplemented with vitamin D through a tube. Ten days later he ended up in the neonatal intensive care unit and tested positive for quite a few forms of bacteria. He stopped breathing on several occasions but was revived each time and put on a cocktail of antibiotics. He responded favorably to the antibiotics and his parents were later told that he had an E. coli urinary tract infection.
Twelve years later, after being bitten by an insect that presumably also carried bacteria, Matt started to suffer from intense headaches and recurrent infections, including bladder infections. Gradually, he developed full-fledged chronic Lyme disease. It is very likely that the pathogens he had acquired at birth had persisted in his tissues, and accompanied by the bacteria introduced by the insect bite, caused him to develop a serious chronic disease later in life.
“We have always wondered why, out of the 1,600 boys in Matt’s school, he is the only one in recent memory who has missed a couple of years of schooling due to a chronic disease. We are sure he is not the only one to be bitten by an insect,” says Matt’s mother, Robyn Russell.
Airborne dispersal
In 2011, Wilkinson et al. used large computer models of the Earth's atmosphere to research how widely microbes could be dispersed, determining that once the microbes of 0.02mm in diameter and below were airborne, they can easily travel thousands of kilometers.
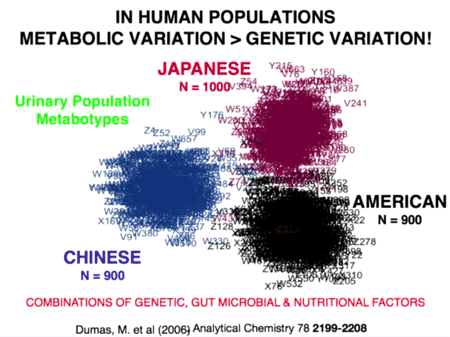
Metabolic evidence for transmission of bacteria
In 2006, Jeremy Dumas et al analyzed the non-human metabolites in urine taken from population samples in Aito Town, Japan; Chicago, IL; and Guangxi, China.55) The team plotted the differences between those metabolites on a two-dimensional chart (right). As is plainly evident, the non-human metabolites from each region are distinct.
Interestingly, people who recently moved between regions began to take on the metabolic profile of that region. One particular artifact is that there are a number of violet numbers in the American group. These represent five Japanese men and women who recently moved to America. When they did that, their metabalomes suddenly took on the American characteristics rather than their native Japanese characteristics.
Dumas's work shows that the makeup of the “metabolome” owes more to environmental factors than genetic ones. To this end, a 2004 study by Wirth showed that two human ethnic groups based in India, which could not be distinguished on the basis of human DNA markers, could be distinguished based on their patterns of H. pylori variation.56)
Disabled immune response increases susceptibility to acute infections
Certainly the exposure to and acquisition of new bacteria plays a role in the development of disease, but these factors don't account for everything. Diseases appear to strike randomly if for no other reason than their massive incidence and mortality. But, a close look at the evidence suggests that diseases tend to strike those who are most vulnerable. This is true even for acute infections such as the Black Death, the deadliest known epidemic in human history. The results of one analysis of 14th century skeletal remains found that “the Black Death did not kill indiscriminately - that it was, in fact, selective with respect to frailty….”57)
During the SARS epidemic, those who were succumbing were people with weakened immune systems, especially diabetics and healthcare workers.58) With the recent Escherichia coli O157:H7 epidemic, there appears to be a demographic pattern of patients who are likely already heavy carriers of a pathogenic microbiotaThe bacterial community which causes chronic diseases - one which almost certainly includes multiple species and bacterial forms.. Remember that, like SARS, a lot more people are getting infected than those who actually fall ill and can't recover.
Trevor Marshall, PhD
According to the Marshall PathogenesisA description for how chronic inflammatory diseases originate and develop., “frailty” could just as easily refer to the strength of immune response. It is the absence of a robust immune response which is the primary contributing determinant in whether someone gets sick with chronic illness or someone remains healthy.
Bacterial pathogens themselves can make a human host more hospitable to their growth and reproduction by secreting substances like the sulfonolipid capnine, which bind and block the Vitamin D ReceptorA nuclear receptor located throughout the body that plays a key role in the innate immune response., a nuclear receptorIntracellular receptor proteins that bind to hydrophobic signal molecules (such as steroid and thyroid hormones) or intracellular metabolites and are thus activated to bind to specific DNA sequences which affects transcription. that controls the innate immune response. Over time, bacteria succeed in suppressing the immune response through a gradual process known as successive infection. In the absence of intervention, successive infectionAn infectious cascade of pathogens in which initial infectious agents slow the immune response and make it easier for subsequent infections to proliferate. is something of an inevitability as everyone who lives long enough will take on the aches, pains, memory loss, and other symptoms that are the hallmark of chronic disease.
Bacteria are allowed to further proliferate when a person consumes any number of immunosuppressive foods, drugs, supplements and other substances. These substances include: immunosuppressants, beta-lactam antibiotics such as penicillin, corticosteroids, and foods and supplements containing high levels of vitamin D. The consumption of such substances is at historical levels and may be largely responsible for the recent spike in chronic disease incidence.
Read more
- Black Death study lets rats off the hook – The Plague of 1348-49 spread so fast in London the carriers had to be humans not black rats, says archaeologist.
- Colic may be caused by H. pylori – Colic is a condition in which an otherwise healthy baby cries for more than three hours per day, more than 3 days per week. A 2012 Egyptian study used a stool antigen test to determine if the babies were infected if infants had Helicobacter pylori. Among the 55 infants with colic, 45 (81.8%) tested positive for H. pylori. In the control group, only 7% of the babies tested positive for H. pylori.59)
- Farm Use of Antibiotics Defies Scrutiny – A dearth of information makes it difficult to document the precise relationship between routine antibiotic use in animals and antibiotic-resistant infections in people, scientists say.
[PMID: 19098867] [DOI: 10.1038/ajg.2008.61]
[PMID: 8554430] [DOI: 10.3109/01485019508987852]
[PMID: 12447413] [DOI: 10.1038/420265a]
[PMID: 19948036] [PMCID: 2794269] [DOI: 10.1186/1477-7827-7-140]
[PMID: 22404291] [DOI: 10.1111/j.1365-2605.2012.01256.x]
[PMID: 18971361] [PMCID: 2620857] [DOI: 10.1128/JCM.01206-08]
[PMID: 18725970] [PMCID: 2516597] [DOI: 10.1371/journal.pone.0003056]
[PMID: 1437883] [DOI: 10.3109/15513819209024224]
[PMID: 7884221] [DOI: 10.1016/s0163-4453(94)91174-6]
[PMID: 17888583] [DOI: 10.1016/j.mehy.2007.06.012]
[PMID: 15482749] [DOI: 10.1016/j.fertnstert.2004.05.076]
[PMID: 18928978] [PMCID: 3939789] [DOI: 10.1016/j.ajog.2008.06.040]
[PMID: 17905166] [PMCID: 7159293] [DOI: 10.1016/S0140-6736(07)61515-3]
[PMID: 21041526] [PMCID: 3036184] [DOI: 10.1113/jphysiol.2010.197764]
[PMID: 16836820] [PMCID: 3291059] [DOI: 10.3201/eid1207.060037]
[PMID: 17594176] [PMCID: 1896187] [DOI: 10.1371/journal.pbio.0050177]
[PMID: 18449059] [DOI: 10.1097/INF.0b013e318169ef47]
[PMID: 16604192] [PMCID: 1430358] [DOI: 10.1172/JCI27372]
[PMID: 12438429] [PMCID: 2193991] [DOI: 10.1084/jem.20020943]
[PMID: 20577268] [PMCID: 3899649] [DOI: 10.1038/nri2802]
[PMID: 16179970] [PMCID: 2315785] [DOI: 10.1007/s00018-005-5260-7]
[PMID: 12538777] [DOI: 10.1203/01.PDR.0000047471.47777.B0]
[PMID: 17928596] [DOI: 10.1056/NEJMoa052632]
[PMID: 17928604] [DOI: 10.1056/NEJMe078119]
[PMID: 17684288] [DOI: 10.1513/pats.200607-138MS]
[PMID: 9830183] [PMCID: 1313221]
[PMID: 16844727] [PMCID: 2121155] [DOI: 10.1136/thx.2006.062836]
[PMID: 8214953] [DOI: 10.1164/ajrccm/148.4_Pt_1.974]
[PMID: 17652652] [DOI: 10.1056/NEJMsa066082]
[PMID: 22685148] [PMCID: 3406147] [DOI: 10.1128/AEM.01361-12]
[PMID: 22920043] [PMCID: 3490988] [DOI: 10.1186/1471-2180-12-184]
[PMID: 21498385] [PMCID: 3079400] [DOI: 10.1093/cid/cir181]
[PMID: 22004107] [DOI: 10.1111/j.1462-2920.2011.02614.x]
[PMID: 22875910] [DOI: 10.1136/bjsports-2012-091395]
[PMID: 17560693] [DOI: 10.1016/j.vaccine.2007.03.053]
[PMID: 19033158] [DOI: 10.1093/aje/kwn310]
[PMID: 15562126] [DOI: 10.1001/jama.292.20.2478]
[PMID: 15317028] [DOI: 10.1002/pds.998]
[PMID: 14985487] [DOI: 10.1056/NEJMoa030595]
[PMID: 8183577]
[PMID: 12459734] [DOI: 10.1038/nrc947]
[PMID: 11205211]
[PMID: 16027303] [DOI: 10.1001/archderm.141.7.869]
[PMID: 18258148] [DOI: 10.2310/7750.2007.00040]
[PMID: 12002380]
[PMID: 12172448] [DOI: 10.1097/00063198-200209000-00015]
[PMID: 18337601] [PMCID: 2361129] [DOI: 10.1056/NEJMoa074943]
[PMID: 9574684] [PMCID: 104807] [DOI: 10.1128/JCM.36.5.1240-1244.1998]
[PMID: 15050933] [DOI: 10.1016/S1473-3099(04)00965-X]
[PMID: 16579598] [PMCID: 6561113] [DOI: 10.1021/ac0517085]
[PMID: 15051885] [PMCID: 387319] [DOI: 10.1073/pnas.0306629101]
[PMID: 18227518] [PMCID: 2234162] [DOI: 10.1073/pnas.0705460105]
[PMID: 16172857] [PMCID: 7088345] [DOI: 10.1007/s10096-005-0004-z]
[PMID: 22751879] [DOI: 10.1001/archpediatrics.2011.1241]