Related article: Koch's postulates

Table of Contents
Detecting bacteria
It has been called one of the greatest false prophecies in the history of medicine. In 1821, Sir John Forbes concluded that the newly invented stethoscope was “ludicrous” and would never be generally adopted by physicians.1) However, this honor might better be bestowed upon many members of the microbiology community, who before the advent of molecular technology, deemed the body to be largely sterile.
Until very recently, efforts to detect and identify microorganisms have depended on in vitroA technique of performing a given procedure in a controlled environment outside of a living organism - usually a laboratory. studies – research in which bacteria were grown in culture in a laboratory setting. As a result, many researchers began to assume that chronic diseases were not caused by microbes. The net effect of all this was that the understanding of pathogens in disease was driven by the study of well-known, easy-to-culture microbes – which, as it turns out, represents the vast minority of bacteria in the human body. By one estimate, 99.6% of the species in the human microbiotaThe bacterial community in the human body. Many species in the microbiota contribute to the development of chronic disease. have not or cannot be characterized through culturing techniques.
As an alternative to traditional methods for culturing, various in silicoExperiment technique performed on computer or via computer emulation. methods for DNA and RNA sequencing have been developed including polymerase chain reaction (PCR). Recent experiments using PCR have offered compelling proof that traditional cultivation methods offer only a limited glimpse into the full extent of the human microbiota and that those microbes found play a role in disease.
Cultivating bacteria in a laboratory setting
Until very recently, efforts to detect and identify microorganisms have depended on in vitro studies – research in which bacteria were grown in culture in a laboratory setting. Because at least a fraction of microorganisms are not particular in their growth requirements, these efforts have yielded an array of diverse microbial cultivation techniques. Microbial cultivation methods opened up an unsuspected world of microscopic life and presumed causative agents of human illness.2)
Koch's postulatesCentury-old criteria designed to establish a causal relationship between a causative microbe and a disease. Koch's belief that only one pathogen causes one disease has now been called into question as multiple postulates are increasingly considered out of date. is a set of ground rules to determine whether a given organism can cause a given disease. One of its dictates is that a bacterium must be shown to grow outside the body in culture in order to prove that the bacterium causes disease. For at least a century, Koch's postulates have held sway.
According to Robert KochAuthor of Koch's postulates, a set of rules for establishing a relationship between a causative microbe and a disease. Koch's belief that only one pathogens causes one disease has now been called into question as multiple postulates are increasingly considered out of date., or at least the ideas since attributed to him, there were no cultivation-resistant microbes. Over the years, varied researchers had difficulty consistently culturing bacteria found in disease. As a result, many researchers began to assume that chronic diseases were not caused by microbes. The net effect of all this was that the understanding of pathogens in disease was driven by the study of well-known, easy-to-culture microbes3) – which, as it turns out, represents the vast minority of bacteria in the human body.
A history of discounting culture-independent methods of microbe identification
The Wirostkos were Columbia University-based opthamologists who used scanning electron microscopy to document the existence of mycoplasma-like organisms in the leukocytes of the eyes. These organisms occurred in inflammatory areas in several conditions: uveitis, vitritis, ocular disease in juvenile rheumatoid arthritis, Crohn’s disease, and sarcoidosis. However, this body of work has been disregarded largely because of an absence of culture-based techniques, as a 1990 Lancet editorial complains:
The case for uncultured organisms even existing let alone remaining uncultured has not been proven and repetitive uncontrolled observations do not serve the scientific community well and may mislead the unwary.
Anonymous Lancet editorial 4)
More than two decades later, it remains unclear the full extent to which this presumption remains in place. What is clear is that this presumption has sidetracked a lot of fruitful scientific inquiries.
The Great Plate Count Anomaly
Culturing always favors the recovery of organisms that are best able to thrive under laboratory conditions (colloquially “lab weeds”), not necessarily the dominant or most influential organisms in the environment.
National Research Council, 2007 5)
Researchers have long known that traditional methods for culturing bacteria are effective at identifying only a fraction of the bacteria in a given sample. One of the first reports of this came from Razumov6) who noted in 1932 that a large discrepancy between the viable plate count and total direct microscopic count of bacteria taken from aquatic habitats. Razumov found higher numbers (by several orders of magnitude) by direct microscopic counting than by the plating procedure.
In 1949, Winogradsky confirmed Razumov's assessment also noted that many microbes are not satisfied with laboratory cultivation conditions. He remarked that readily cultivated bacteria in natural microbial communities “draw importance to themselves, whereas the other forms, being less docile, or even resistant, escape attention.”7)
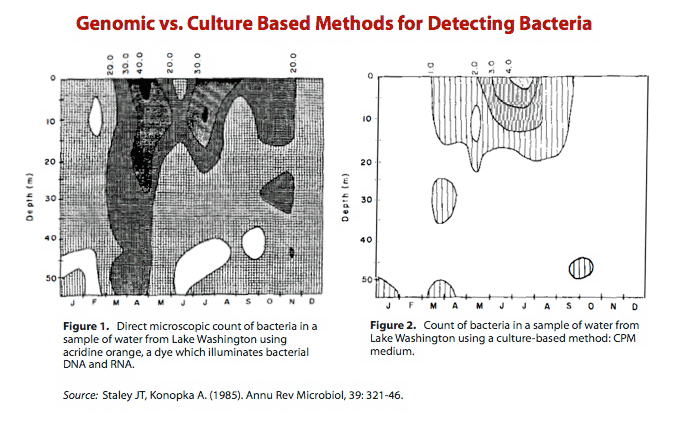
In 1985, Staley and Konopka pointed to Razumov's discrepancy and called it “the Great Plate Count Anomaly.”8) Their review describes work in which the researchers took hundreds, perhaps even thousands, of water samples from Lake Washington and performed two methods for counting bacteria.
Figure 1 shows the number of bacteria identified using a fluorescent dye, acridine orange. Acridine orange counts bacteria by interacting with bacterial DNA and RNA.
Figure 2 shows the number of bacteria identified in a culture-based medium – essentially a Petri dish.
As it is plainly evident, the genomic method for detecting bacteria (Figure 1) was orders of magnitude more sensitive than a method based on viability in a culture (Figure 2). The shortcomings of the cultivation method is striking and suggests that this traditional method for cultivation is only effective in identifying a fraction of all bacteria.
The author's conclusions have been used even very recently to apply to cells everywhere including in the human body:9)
As the figures illustrate, only approximately 0.1-1.0% of the total bacteria can be enumerated by the plating procedure. Indeed, as a general rule we have found that the maximum recovery of heterotrophic bacteria [bacteria that don't use photosynthesis] is 1% of the total direct count using plating procedures or other viable enumeration methods…. From a microbiological perspective, only a few percent of the bacterial cells enumerated by direct microscopic count can be cultured and identified (see previous section on plate counts). No breakthrough in determining species diversity seems likely in the near future.
James T. Staley and Allan Konopka, PhD 10)
Over the years, researchers have pointed out two reasons why the majority of bacteria that comprise the human microbiomeThe bacterial community in the human body. Many species in the microbiota contribute to the development of chronic disease. do not culture:
- Some forms and kinds of bacteria only grow in specific conditions offered by the human body including a very narrow pH, the right nutrient availability, etc. For example, intracellular bacteria grow poorly or not at all on stand culture media.11)
- Certain bacteria only grow in the presence of certain other species of bacteria.
By David Relman's estimate, 99.6% of the species in the human microbiota have not or cannot be characterized through culturing techniques.12)
According to a 2009 review by James D. Oliver, “The debate over whether a VBNC [viable but not culturable] state truly exists has largely been put to rest, largely as a result of numerous molecular studies reported in recent.”13) Oliver catalogs the various factors, both chemical and environmental shown to induce the viable but not culturable state:
- nutrient starvation
- incubation outside the normal temperature range of growth
- elevated or lowered osmotic concentrations
- commonly used food preservatives
- heavy metals
- exposure to white light
The switch to the VBNC stage has been described and documented for at least the following bacterial species: Vibrio spp. (cholerae, vulnificus and other species), Escherichia coli (including EHEC), Campylobacter jejuni, Helicobacter pylori, Salmonella spp., Listeria monocytogenes, Yersinia enterocolytica, Shigella spp., Klebsiella spp., Enterobacter spp., Cronobacter spp., Staphylococcus aureus, Providencia spp., Morganella spp., Pseudomonas spp., Mycobacterium tuberculosis, Enterococcus spp. A number of studies have shown that processes meant to achieve bactericidal effects can favor bacterial switch to VBNC.14)
DNA and RNA amplification techniques
As an alternative to traditional methods for culturing, various in silico methods for DNA and RNA sequencing have been developed. The most commonly used DNA and RNA amplification techniques is polymerase chain reaction (PCR), a technique to amplify a single or few copies of a piece of DNA across several orders of magnitude, generating millions or more copies of a particular DNA sequence
One commonly used practice is to look at a bacterial species' 16S RNA, a sequence of genetic code which has the benefits of both being conserved across bacterial species15) and a historical tendency to change at a regular, but limited, rate over time over long periods of time.
Currently, the conventional 16S rDNA PCR technique must involve three steps. First, a step that corresponds to the amplification and revelation of amplified products on agarose gels. Second, a step in which the amplified product is sequenced. Third, a step in which the obtained sequence is analyzed and compared with that given in a database, mainly the GenBank, containing all known bacterial sequences to allow an accurate identification. A sequence similarity of less than 97% of the 16S rRNA sequence is the criterion used to define a potentially new bacterial species.16)
Need for improvement
Polymerase chain reaction is a relatively specific and sensitive test, however:
As we discuss in our new book chapter, PCR is of little use when examining components of the microbiotaThe bacterial community which causes chronic diseases - one which almost certainly includes multiple species and bacterial forms., as bugs adjust their genomes when they go into the microbiota, and the specific fragments of the organism, for which conventional PCR tests are looking, may not be present while the microbe is a member of the microbiota.
On resurgence to an acute-phase pathogen, they get those genes back, by methods not really fully understood.
Trevor Marshall, PhD
Another problem is that PCR primers may miss half of rRNA microbial diversity17) According to Hong et al.'s work, “even with unlimited sampling and sequencing effort, a single combination of PCR primers/DNA extraction technique enables theoretical recovery of only half of the richness recoverable with three such combinations.”
Existence of microbes in healthy tissue supposed to be sterile
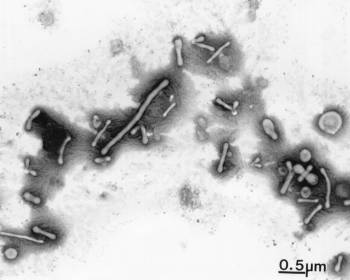
- blood – The presence of microbes in the blood is known as bacteremia. While many doctors believe it to be rare, a significant blood microbiome is ubiquitous. The Relman Lab at Stanford used real time PCR to show that there is a substantial and “normal” population of bacterial DNA sequences in the blood of even healthy individuals.18) McLaughlin et al. used 16S rRNA probes and dark field microscopy to show that the blood of healthy individuals contained pleomorphic microorganisms, which grow slowly and are not susceptible to antibiotics (right).19) They concluded that their findings “should not be controversial.” This work has been since confirmed by Moriyama.20)
- lungs – Erb-Downward found bacterial 16S sequences in the lung tissues of all subjects in a 2011 based study of healthy patients as well as those with chronic obstructive pulmonary disease (COPD).21)
Role of cultivation-resistant microbes in disease
Regardless of the role that the VBNC [viable but not culturable] state plays, it is clear that a large number of non-spore-forming bacteria, most notably a large number of human pathogens, are capable of entering this state, maintaining cellular structure and biology and continuing significant gene expression while otherwise nonculturable by “standard” laboratory methods. That they can exit from this state, and become culturable again, is also undeniable. Finally, it can no longer be questioned that the VBNC state plays a critical role in the survival of important human (and other) pathogens, and possibly in their ability to produce disease.
James D. Oliver 22)
Recent experiments using PCR have offered compelling proof that traditional cultivation methods offer only a limited glimpse into the full extent of the human microbiota and that those microbes found play a role in disease.
- amniotic fluid – One study compared the rate of amniotic fluid infection with pre-term delivery among pregnant women (see right).23) The main conclusion of the study was that the presence of infection is correlated with pre-term delivery. But, perhaps the most interesting outcome of the research related to the techniques for determining infection. In a cohort of 166 who went into preterm labor, 19 subjects were found to be positive for infection using a molecularly-based genetic sequencing technique called PCR, including 9 that were missed with a traditional culture. In fact, the positive predictive value of PCR for preterm delivery was 100 percent. The PCR technique picked up on the presence of 18 different taxa of bacteria as opposed to 11 taxa using conventional cultivation techniques.
- chronic wounds – Another study examined the bacteria present in three types of chronic wounds: diabetic foot ulcers, venous leg ulcers, and pressure ulcers.24) Researchers used both traditional cultivation and a battery of molecular techniques for identifying bacteria. In only one wound type did culture methods correctly identify the primary bacterial population.
- urinary tract infections –
Urinary tract infections (UTIs), the most common kidney and urologic diseases in industrial nations, are usually caused through faecal contamination of the urinary tract. In this study, we have examined 1449 urine specimens both by culture and by PCR. The majority of UTIs examined were caused by Escherichia coli (35.15%), followed by miscellaneous bacteria (23.03%), and by Enterococcus faecalis (19.39%). A large fraction of fastidious and anaerobic bacteria (22.43%) was not detected under culture conditions but only by using PCR. This group of bacteria evade the standard culture conditions used in routine diagnostic laboratories examining urine specimens. The molecular approach used broad-range 16S rDNA PCR, denaturing high-performance liquid chromatography analysis, sequencing, and bioinformatic analysis to uncover these 'hidden' pathogens and is recommended in particular when examining leukocyte esterase-positive and culture-negative urinary tract specimens.
Pervin Imirzalioglu et al.25)
- atherosclerosis – Sometimes novel uses of culture-based techniques can confirm the presence of additional microbes. Using an array of new culture-based techniques, Kozarov et al. recovered isolates from Propionibacterium acnes, Staphylococcus epidermidis and Streptococcus infantis and the fastidious anaerobe Porphyromonas gingivalis. In fact, 3,500 genomes of P. gingivalis were found per gram of atheromatous tissue.26)
Availability of tests influence which microbes are studied in chronic diseases
Certain pathogens are repeatedly linked to various inflammatory conditions, but this does not mean these pathogens are the only microbes influencing the disease state. What these results may largely imply is that scientists have created effective tests that easily pick up on the presence of these microbes. Other microbes that may also be causing disease do not have a standard laboratory test to detect their presence.
H. pylori is often associated with a number of stomach conditions, because there is a reliable easy-to-use test to detect its presence, however, there are hundreds of other pathogens in the gut capable of causing disease that are not able to be detect in a standard laboratory. So, the microbe that ends up being discussed most in connection to stomach diseases is H. pylori, largely because the availability of existing tests.
As tests for communities of bacteria are developed and become more widely available, the understanding of microbes' effects on disease should evolve.
[PMID: 7026784] [PMCID: 1439259]
[PMID: 9716951] [PMCID: 2640285] [DOI: 10.3201/eid0403.980310]
[PMID: 19040704] [DOI: 10.1111/j.1365-2672.2008.03981.x]
[PMID: 1970375]
[PMID: 18502944] [PMCID: 2493393] [DOI: 10.1101/gr.075549.107]
[PMID: 3904603] [DOI: 10.1146/annurev.mi.39.100185.001541]
[PMID: 20586563] [DOI: 10.1586/eri.10.52]
[PMID: 20059548] [DOI: 10.1111/j.1574-6976.2009.00200.x]
[PMID: 21038700]
[PMID: 14612235] [DOI: 10.1016/S0378-1097(03)00717-1]
[PMID: 18525362] [DOI: 10.1097/BOR.0b013e3283032030]
[PMID: 19693101] [DOI: 10.1038/ismej.2009.89]
[PMID: 11326021] [PMCID: 88056] [DOI: 10.1128/JCM.39.5.1956-1959.2001]
[PMID: 12454193] [PMCID: 154583] [DOI: 10.1128/JCM.40.12.4771-4775.2002]
[PMID: 18667036] [DOI: 10.1111/j.1348-0421.2008.00048.x]
[PMID: 21364979] [PMCID: 3043049] [DOI: 10.1371/journal.pone.0016384]
[PMID: 18725970] [PMCID: 2516597] [DOI: 10.1371/journal.pone.0003056]
[PMID: 18325110] [PMCID: 2289825] [DOI: 10.1186/1471-2180-8-43]
[PMID: 18336452] [DOI: 10.1111/j.1439-0272.2007.00830.x]
[PMID: 21366733] [PMCID: 3133811] [DOI: 10.1111/j.1365-2796.2011.02373.x]